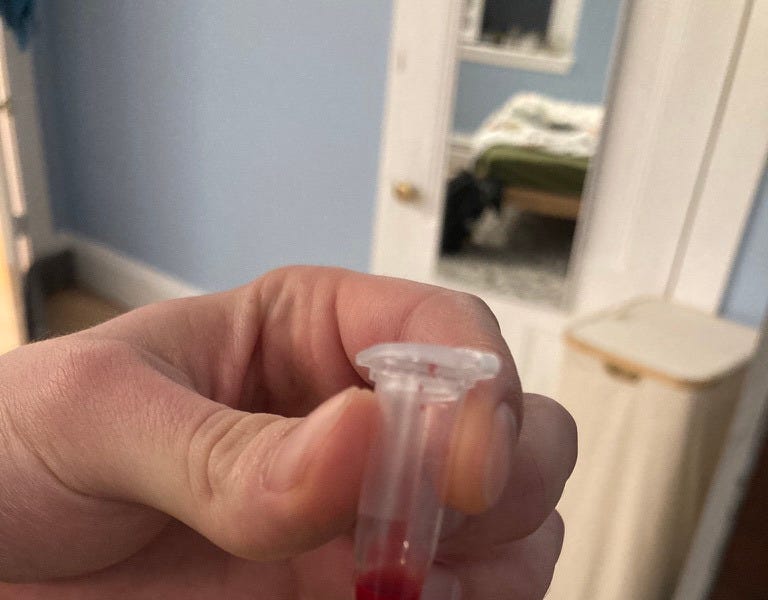
“It really hurts”
My finger drips with blood.
“God… do you want me to do it for you?”
“Sure, I guess”Before I can say anything more, she stabs the needle into my finger again. And again. And again.
More rivulets of blood along my pinky, my eyes tear up, and she squeezes my finger over the test tube.
“OK, you can go clean up.”
The first human genome took $2.3 billion dollars and 13 years to sequence.
Today, you can use an Oxford Nanopore ($1000 dollars) to sequence whatever DNA you want in less than 48 hours. Or so their marketing materials say.
Right now, if you want to sequence your own DNA, you have to send blood or a cheek swab to some shady third party that, for all I know, gives my DNA away on a USB stick with 10,000 other peoples’ DNA in a dark corner of Shenzhen.
Some friends and I became curious: How hard would it be for us to buy a Nanopore and sequence DNA, using makeshift equipment in our bedroom?
The goal is to get from 10ml of blood to a string of 3 billion A’s, C’s, G’s, and T’s.
The rough steps are pretty straightforward:
-
Get blood sample
-
Extract human DNA from all the other goop
-
Run the DNA through small electric holes so the Nanopore can read individual base pairs of each DNA fragment
Before getting into the equipment, please allow me a quick technical digression into how DNA sequencing has changed over the years:
A summary of the Three Ages of DNA Sequencing from this excellent paper.
-
Sanger Era: Very Slow (1960-2003)
Sanger et al. figured out you could stop DNA polymerase at specific base pairs by feeding it defective nucleotides. Add a bunch of defective nucleotides and this happens more or less at random, resulting in a bunch of DNA strands at different lengths.
Add an electric current across a gel and the smaller bits will separate. You can then read the bands like a barcode.
This approach took 13 years and $2.3 bn to do the human genome (~3 billion bp).
-
Illumina Era: Parallelization (2005-2010s)
‘Sequencing by synthesis’. instead of chopping up and separating each base pair through a gel lattice, we
-
Single molecule sequencing
Now we use electrical nanopores and read the individual charge of each base pair. Nothing needs to get chopped up, the processed DNA is unwound and passed through a tiny hole. each base pair like beads on a molecular string pulled through a hole
This is the approach we used.
-
Oxford Nanopore MinION starter kit ($1,000) – includes the USB sequencer, flow cell with tiny biological pores, and prep chemicals
-
Zymo DNA extraction kit (free sample from their website)
-
Mini centrifuge from Amazon ($50) – spins stuff fast
-
lab consumables: eppendorf tubes, lancets, pipette ($50)
Total cost: ~$1,100

We had to collect 200 µL of blood (about ⅕ of a ml).
I made a bad mistake by buying diabetes lancets that were too small, and each prick only yielded about 10 µL. It was unsurprisingly horrible to stab my finger again and again so Cece kindly took the lancet and rapidly jabbed it into my pinky until there were multiple tiny beads we could squeeze out.

Blood itself has all this gunk in it – mostly red blood cells, which don’t even have DNA. You need to dissolve the white blood cell membranes and separate out this DNA. The Zymo kit gave us the right enzymes and a special test tube that acted as a filter when spun through a centrifuge. We followed the instructions using our thermal cycler which worked fine.
We then had to follow the Nanopore prep kit, which basically had us attach adapter molecules so the nanopore could tell what it was reading.

We pipetted the prepared DNA into a tiny port on the MinION flow cell – a device smaller than a pencil case. We plugged the whole thing into a USB port, fired up the MinKNOW software, and watched in real-time as the software did ‘basecalling’ — as far as I can tell it feeds electrical signals into a neural net and predicts the signature as an A, T, C or G in real time.


So I have to acknowledge that the results were basically useless for analysis.
We sequenced around 1 gigabase total across two runs (the first died during a hardware error). The human genome is around 3 gigabase so we got about 13% coverage. Another problem is that 75% of what we sequenced was actually human DNA – the other 25% was some form of bacterial contamination.
I wanted to look at my single nucleotide polymorphisms (eg lactose tolerance genes, breast cancer genes). To confidently call SNPs you want to sequence the same section multiple times. We had most spots sequenced zero or one times.
Another problem was our flow cell was malfunctioning from the start — only 623 out of 2048 pores were working.
Still! We sequenced a meaningful fraction of my human genome for $1100.
Acknowledgements
Thank you to my dear friends Lenni, Cece, and S for conducting this experiment with me